Day 7 Handy Haversacks
This is my attempt to solve Day 7.
7.1 Part 1
I think today’s problem is naturally solved with using graph’s, so I’m going to use the {igraph} package. The easiest
way to create a graph is to first create a data frame that contains the columns from and to that indicate which
edges are in the graph. Any additional columns will be added as properties to the edge.
convert_input_to_tibble <- function(input) {
input %>%
# we don't need to keep the word's "bag" or "bags". In our data these words
# always have a space before, and sometimes have a "." at the end. We can
# use the following regex to remove these words
str_remove_all(" bags?\\.?") %>%
# we can now split our data at the word "contains"
unglue_data("{from} contain {contains}") %>%
# now we can split the contains column by comma's
mutate(across(contains, str_split, ", ")) %>%
# and expand the nested "contains" column
unnest_longer(contains) %>%
# handle edge case: if the string is no other need to insert a 0 at the start
mutate(across(contains, ~if_else(.x == "no other", "0 no other", .x))) %>%
# now we can split the contains column into the "n" part and the "to" part
separate(contains, c("n", "to"), "(?<=\\d) ", convert = TRUE) %>%
# reorder the columns
select(from, to, n)
}
sample_df <- convert_input_to_tibble(sample)
actual_df <- convert_input_to_tibble(actual)
sample_df## # A tibble: 15 x 3
## from to n
## <chr> <chr> <int>
## 1 light red bright white 1
## 2 light red muted yellow 2
## 3 dark orange bright white 3
## 4 dark orange muted yellow 4
## 5 bright white shiny gold 1
## 6 muted yellow shiny gold 2
## 7 muted yellow faded blue 9
## 8 shiny gold dark olive 1
## 9 shiny gold vibrant plum 2
## 10 dark olive faded blue 3
## 11 dark olive dotted black 4
## 12 vibrant plum faded blue 5
## 13 vibrant plum dotted black 6
## 14 faded blue no other 0
## 15 dotted black no other 0We can create a graph using the graph_from_data_frame() function:
## IGRAPH fd1e49d DN-- 10 15 --
## + attr: name (v/c), n (e/n)
## + edges from fd1e49d (vertex names):
## [1] light red ->bright white light red ->muted yellow
## [3] dark orange ->bright white dark orange ->muted yellow
## [5] bright white->shiny gold muted yellow->shiny gold
## [7] muted yellow->faded blue shiny gold ->dark olive
## [9] shiny gold ->vibrant plum dark olive ->faded blue
## [11] dark olive ->dotted black vibrant plum->faded blue
## [13] vibrant plum->dotted black faded blue ->no other
## [15] dotted black->no otherand visualise this graph using {igraph}:
ggraph(sample_g, layout = "igraph", algorithm = "nicely") +
geom_edge_link(aes(label = n),
angle_calc = "along",
label_dodge = unit(2.5, "mm"),
arrow = arrow(length = unit(3, "mm")),
end_cap = circle(10, "mm")) +
geom_node_label(aes(label = str_replace(name, " ", "\n")))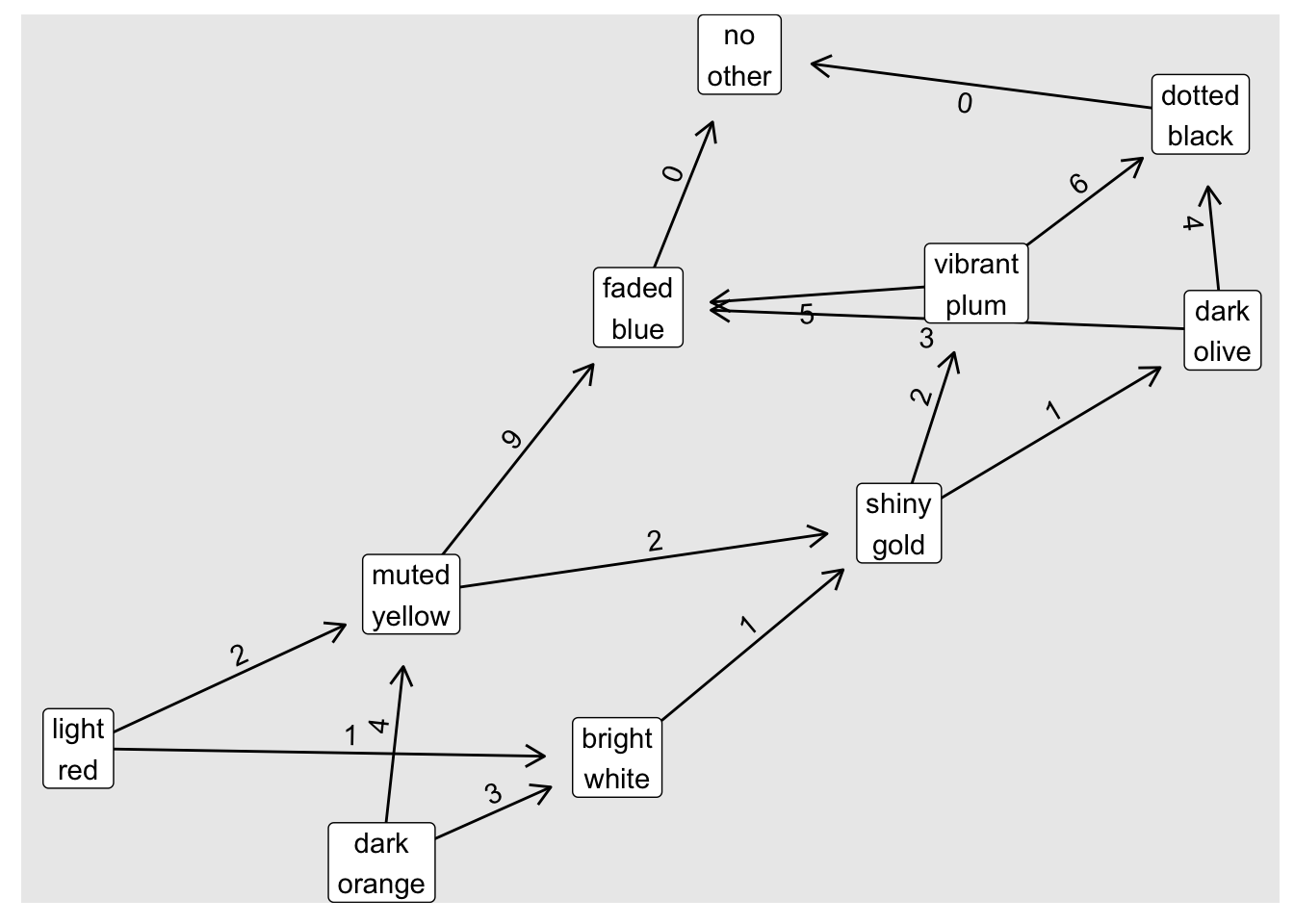
Our challenge is to find all of the vertices that have a path in to “shiny gold”. The subcomponent() function can
tell us all of the vertices in a graph which reach a given vertex. We can simply take the length of this subcomponent
and subtract 1 (as the subcomponent includes “shiny gold”).
## [1] TRUEWe now just need to solve for the actual data.
## [1] 2787.2 Part 2
Part 2 sounds like a recursive function. We can first find the subgraph that includes all of the vertices from “shiny gold”. We can then convert this graph to an adjacency matrix and iterate through each vertex, recursively calling a function that sums how many bags this bag will contain.
part_2 <- function(input) {
sg <- induced_subgraph(input, subcomponent(input, "shiny gold", "out"))
am <- as_adjacency_matrix(sg, attr = "n", sparse = FALSE)
fn <- function(am, v = "shiny gold", n = 1) {
a <- am[v, ] * n
a <- a[a > 0] # keep as separate step, otherwise can end up with just scalar
# now, iterate over each item in a and recursively call this function
sum(map_dbl(names(a), ~fn(am, .x, a[[.x]]))) + sum(a)
}
# call this function on the base case
fn(am)
}We can test that our function works against the sample case:
## [1] TRUEA second sample is provided, so we can test against that also:
c("shiny gold bags contain 2 dark red bags.",
"dark red bags contain 2 dark orange bags.",
"dark orange bags contain 2 dark yellow bags.",
"dark yellow bags contain 2 dark green bags.",
"dark green bags contain 2 dark blue bags.",
"dark blue bags contain 2 dark violet bags.",
"dark violet bags contain no other bags.") %>%
convert_input_to_tibble() %>%
graph_from_data_frame() %>%
part_2() == 126## [1] TRUENow we can run against the actual data:
## [1] 451577.3 Extra: alternative solution to part 2
We could also solve part 2 by iterating through each vertex and finding the incident edges and adjacent vertices. We
multiply the edge weight (ie$n) by the current value of n, sum these values and add n back in to the total. This
gives us the total number of bags, including the initial bag, so we need to subtract 1 from this answer.
part_2_alt <- function(g, v, n) {
ie <- incident_edges(g, v, "out")[[1]]
vn <- ends(g, ie)[, 2]
sum(map2_dbl(vn, ie$n * n, part_2_alt, g = g)) + n
}We can now verify that this alternative function works as above.
## [1] TRUE## [1] TRUEElapsed Time: 2.362s